ROI Buddy GUI¶
Contents
Introduction¶
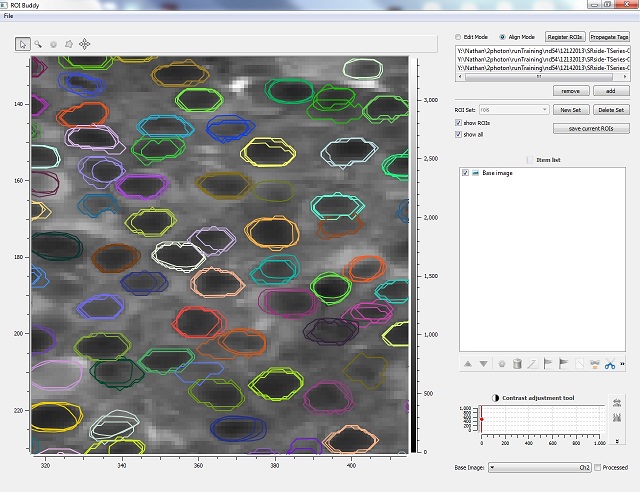
The ROI Buddy GUI can be used for viewing, creating, editing, and tagging the regions of interest (ROIs) associated with multiple SIMA imaging datasets simultaneously. In addition it can be used for registering ROIs from different imaging sessions of the same field-of-view, allowing for the longitudinal tracking of cells in serial imaging experiments.
Typical Workflow¶
- Load multiple SIMA imaging datasets corresponding to different recordings of the same field of view.
- For each dataset, select the channel you wish to edit and align.
- Create or load pre-drawn ROIs for each dataset, editing the ROIs as necessary in Edit Mode.
- Tag cells in one dataset based on some criteria, e.g. morphology.
- Enter Align mode, bringing all the ROIs into alignment.
- Choose “Register ROIs” to assign a common id property to overlapping ROIs.
- Use the merge and unmerge tools to manually fix any incorrect groupings.
- Once registration is satisfactory, choose “Propagate Tags” to assign the morphology tags to all matched ROIs across imaging sessions.
- Save ROIs.
Installation¶
The ROI Buddy GUI is compiled as a Windows executable (.exe) file and is available for download here. Launch the application directly by opening the .exe file.
Alternatively, the ROI Buddy GUI can be built from source. Source code is available for download here. The ROI Buddy GUI depends on SIMA, so shares all dependencies with SIMA. In addition, the following packages are required:
- PyQt4 (http://www.riverbankcomputing.co.uk/software/pyqt)
- guidata (https://code.google.com/p/guidata/)
- guiqwt (https://code.google.com/p/guiqwt/)
Note
The ROI Buddy Windows .exe file compiled with the tifffile module rather than the libtiff C library. If the ROI Buddy GUI is to be used with large TIFF files containing many frames, we recommend running the ROI Buddy from source after installing the libtiff C library and its associated Python bindings, as it enables more efficient memory handling. Alternatively, initializing SIMA iterables with HDF5 datasets enables rapid data access.
Mac OS X¶
We recommend using MacPorts for installing the dependencies. After installing SIMA using MacPorts as described here, run the following command in Terminal to install an additional dependency:
$ sudo port install py27-pyqwt
Then download and install guidata and guiqwt before installing ROIbuddy from the source file.
User Interface¶
Control Panel¶
Toggling modes¶
| Edit Mode | Toggling to edit mode allows for editing, creation, and deletion of ROIs. In addition, this is the mode in which ROI tagging and labeling takes place. |
| Align Mode | Toggling to align mode allows for the transformation of ROI Lists from separate imaging sessions into a common space, allowing for simultaneous visualization of independently drawn sets. In this view, ROI objects can be deleted (as in the case of singleton ROIs that were perhaps a false positive), and ROIs can be registered across days. |
Initializing SIMA imaging dataset and ROI List objects¶
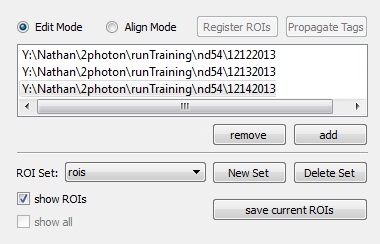
| Remove / Add | Add or remove SIMA datasets from the dataset list. |
| ROI Set | A dropdown allowing for selection of the ROI List currently being modified. These ROI Lists correspond to the keys of imagingDataset.ROIs(). By default, the most recently modified ROI List is displayed upon initialization. |
| New / Delete Set | Create or delete ROI Lists for the active SIMA imaging dataset. These lists are stored in the associated imagingDatset.ROIs dictionary. |
Registering ROI objects across multiple imaging sessions¶
If multiple datasets have been loaded from the same field of view, it is possible to align the ROIs and commonly identify them so that they can be tracked across sessions. The currently selected dataset will act as a template to which all other datasets will be aligned. Each dataset is automatically aligned to the template dataset by calculating an affine transformation between time averaged images that produces maximal similarity. A clustering algorithm based on the Jaccard Index is used to match cells between datasets, which can be manually adjusted by merging/unmerging ROIs from the automatic clusters. Once registered, ROIs that are matched across days are assigned the same id property which is denoted visibly by giving them all the same color.
| Register ROIs | Begin automatic registration using the currently active dataset as the registration template. If datasets do not match, an error dialog will identify the datasets that could be aligned. |
| Propagate Tags | In align mode, the propagate tags button distributes tags across ROI objects from different imaging datasets sharing the same id property. The user can select which tags to propagate. Thus, regions need only be tagged in one dataset, and these tags can be added to other recordings of the same region. |
Warning
In align mode, it is necessary that all imaging datasets loaded must be roughly of the same field of view. Alignment is based upon an affine transformation with 6 degrees of freedom. If a transform between time averaged images cannot be calculated, an error message will be displayed printing the directories of the incompatible sets.
Toggling the visibility of ROIs¶
| Show ROIs / All | Toggle the visibility of ROI polygons. In align mode, Show All results in the transformation of all ROIs into a common space, allowing for visual inspection of alignment across imaging sessions. |
Keyboard shortcuts¶
| m (edit mode): | merge separate ROIs into a single ROI object |
|---|---|
| m (align mode): | merge selected ROIs into the same cluster, assigning them the same ID attribute |
| u (align mode): | unmerge ROI from its cluster and assign it a unique ID attribute |
| f (edit mode): | select freeform tool |
| s (edit mode): | select pointer-selection tool |
| d: | delete |
| r: | randomize ROI colors |